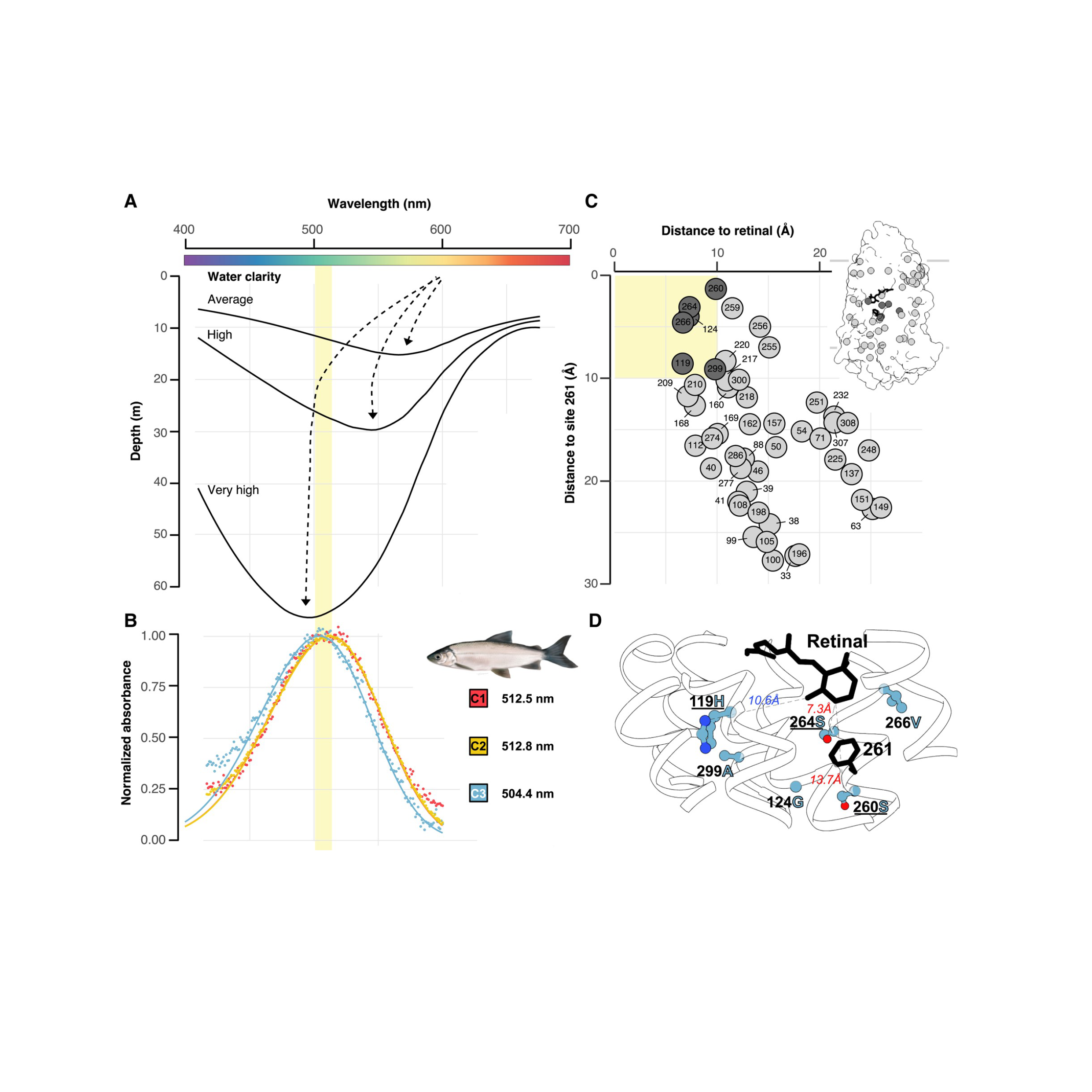

Functional Genomics
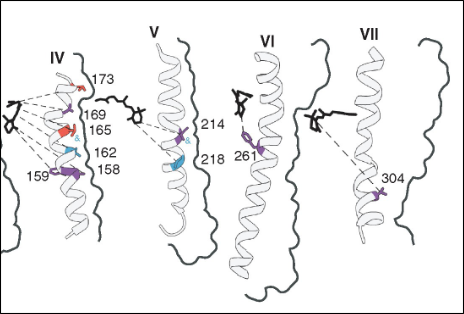
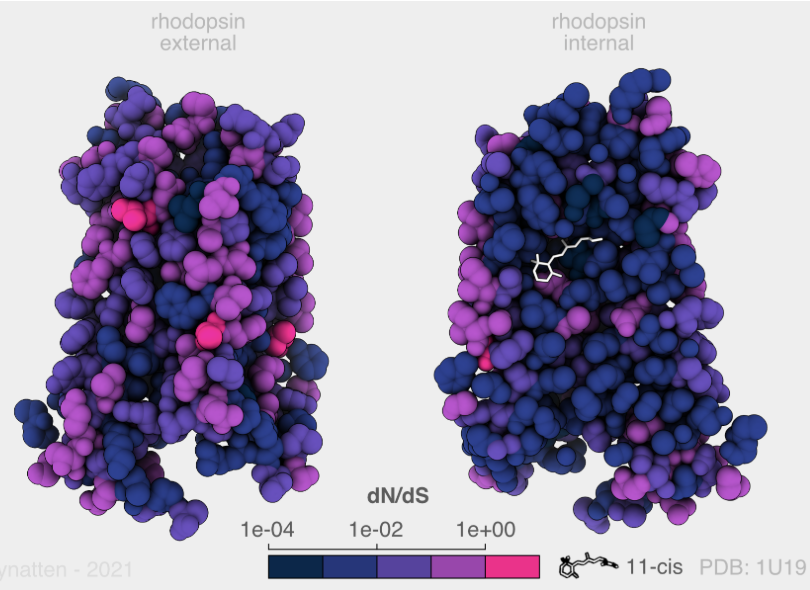
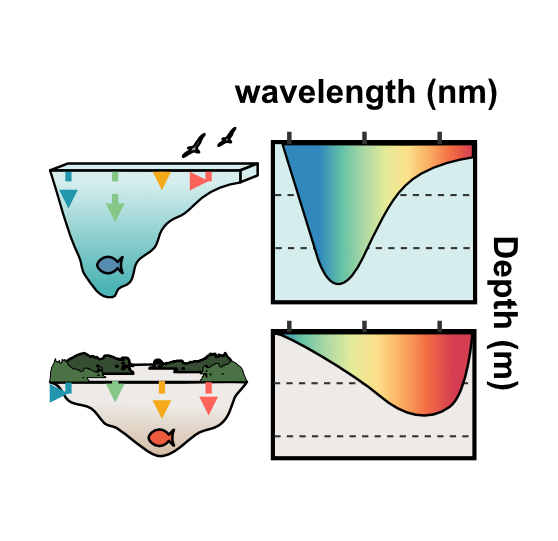
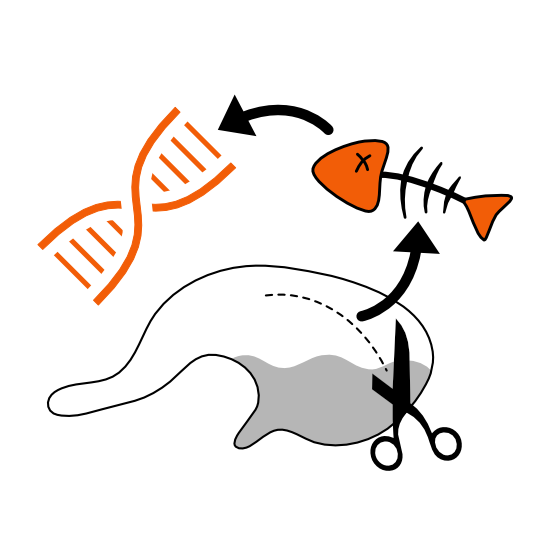
Ecosystem responses to environmental change depend on molecular processes that shape species’ resilience. My research integrates genomics with experimental assays to uncover how environmental differences drive protein evolution. Using visual pigment proteins in fishes as a model system, I have shown how specific amino acid substitutions modulate spectral sensitivity, tuning visual pigments to the spectral conditions of marine and freshwater habitats and maintaining functionally relevant intraspecific genetic variation along depth gradients in Canadian fishes. I have also expanded these approaches to assist with deep mutational scans of visual pigments and functional characterization of other proteins, including arrestins, sodium channels, and CRISPR-associated proteins providing mechanistic insight into the molecular basis of adaptation.

Molecular Ecology
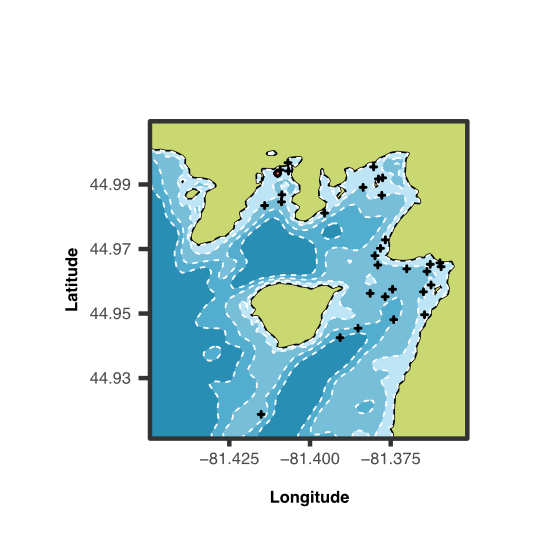
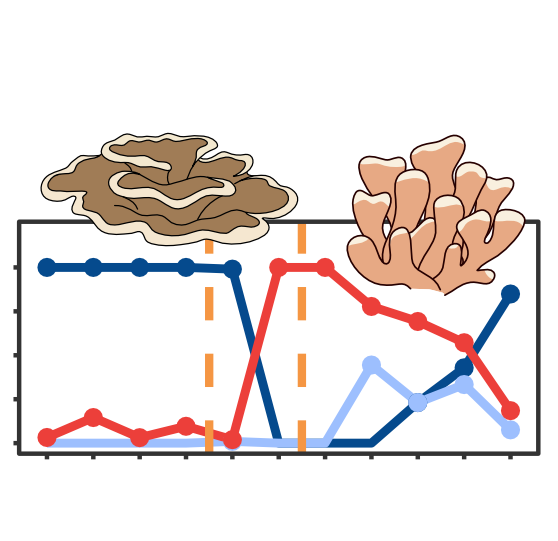
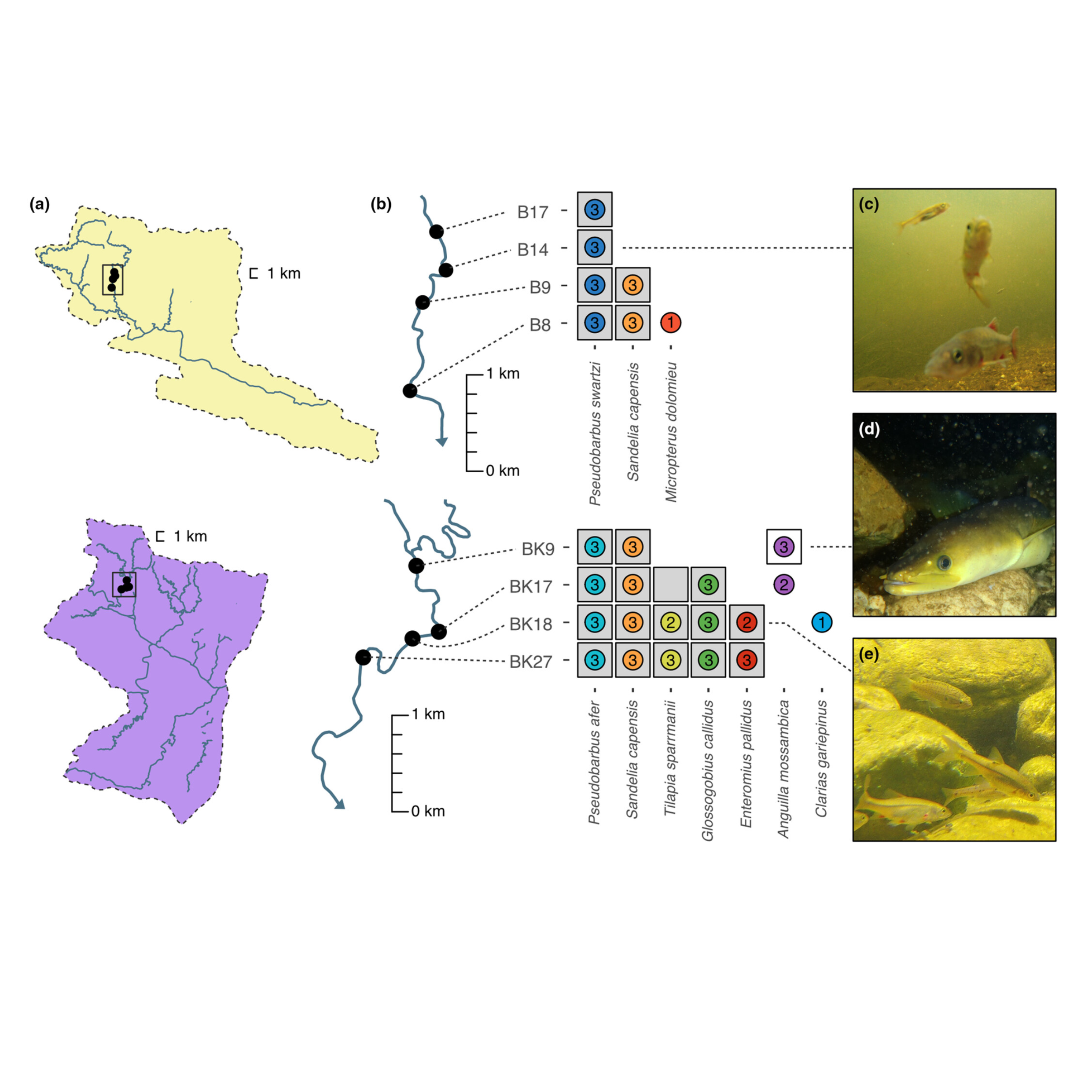
Many analyses of molecular evolution are constrained by limited or incomplete distribution data. I have developed and applied DNA metabarcoding frameworks that link environmental variation to patterns of species and functional diversity across space and time. By integrating data on species presence, reproductive habits, and trophic interactions, my work has advanced how we monitor and interpret ecological change. I have applied these approaches to many systems from sensitive freshwater fish communities in Ontario and South Africa to decade-long surveys of coral reef ecosystems in the Central Pacific, providing new insight into biodiversity responses and resilience to environmental stress. By integrating this work with functional genomics, I am mapping the geographic distribution of adaptive genetic variation linked to protein function and expanding DNA metabarcoding methods to target functionally relevant genes for rapid eDNA surveys across diverse habitats and ecological gradients.